Dissecting Genetic Variability in the COVID-19 Virus
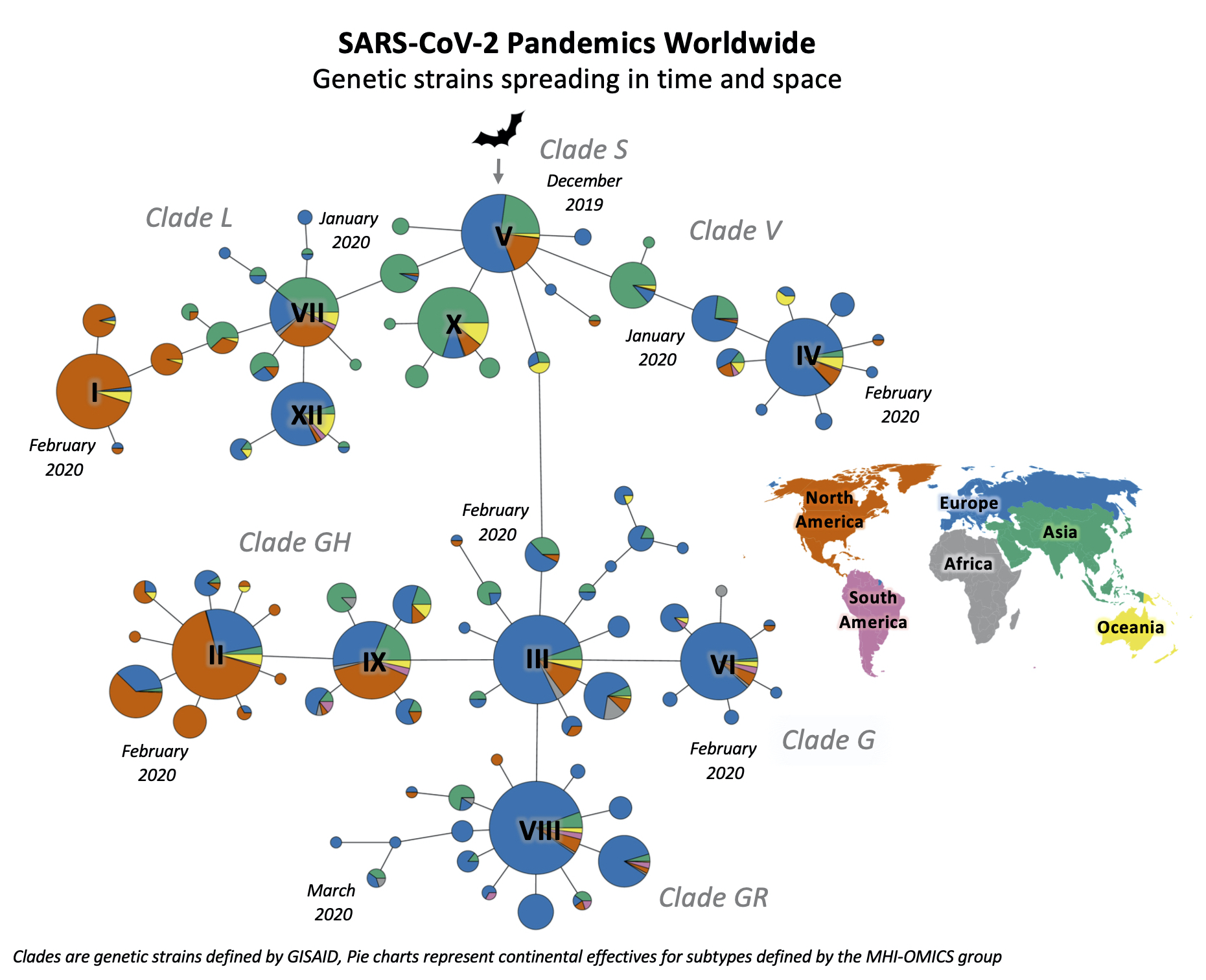
Hussin’s group uses advanced research computing to answer biological questions. These days, they are looking for mutations in SARS-CoV-2, the virus that causes the disease called COVID-19. “We took all the sequences showing up every week and looked at how they mutated from one generation to the next and from one country to another,” says Hussin.

Dr. Julie Hussin, Associate professor, Université de Montréal. Photo crédit : IVADO
“It was only supposed to be a small project with my senior bioinformatician,” says Hussin. While China began reporting huge numbers and cases were showing up in Europe, “we realized that a ton of data on the virus’s genome was emerging and showing variability.” As population geneticists, that variability is their bread and butter.
When the first human was infected with the virus, it had a specific genome. That genome is composed of RNA (whereas the human genome is made of DNA) and codes for everything the virus can do. Upon entering the body, it hijacks the machinery of living cells and turns them into virus factories. When the virus replicates, random errors can slip into the genome. These mutations can be detrimental, beneficial or make no difference to how the virus functions, but they can give rise to new viral strains. Varying pressures inside humans and between people influence which individual viruses survive, replicate and are transmitted. Different strains may each have their own set of advantages.
“The virus’s genome codes specifically for proteins that allow it to enter the cell,” she says. The trick is to figure out which mutations impact its capacity to do that. Which mutations have a strong impact on the viral proteins? Which mutations are interfering with replication or transmission? Which genes need to act together to function?
As COVID-19 spread, the magnitude of what was happening dawned on Hussin’s team. “As scientists, we told ourselves we had to do something with the tools we have, even if we didn’t know if it would work,” she says. “Then we received funding and it just blew up from there,” adds Hussin.
Hussin’s students started applying existing methodologies to study the virus. Her bioinformaticians worked on building “data pipelines,” sequences of data-processing steps, to sort through the raw data.
Bioinformatics is a young and little-known discipline outside of scientific circles. While no-one on her team currently works at a lab bench, their work is key in the development of drug treatments and vaccines against COVID-19. With each coronavirus genome ranging around 30,000 bases in length, Hussin and her programming wizards couldn’t analyze the staggering amount of variation and relationships in the data without Calcul Québec and Compute Canada.
Calculations that take seconds with supercomputers would take hours in her lab’s office desktops. Those that take weeks would otherwise take months or even years. “Without Calcul Québec, we just wouldn’t be able to do this research on time,” she insists.
“We’re still in the collection phase,” says Hussin, “exploring the data as it comes in.” So far, her team has been working on the sequenced genomes of viral samples taken from infected people. What’s missing is the data about those people.
Consortiums of scientists are collecting all available data from those who have tested positive for COVID-19 in Quebec, Canada and around the world. They want to know their medical histories, if they were hospitalized, had complications and, of course, if they survived or not.
Advanced computing along with artificial intelligence tools being developed in partnership with Mila and funded by IVADO—the Institute for Data Valorization—are helping them set the stage for when the human data being collected comes along. Once they start analyzing the viral data alongside the human data, they may be able to identify a subset of patients infected by certain strains of SARS-CoV-2 who will respond to particular treatments.
“Of course,” says Hussin, “we hope there will be a treatment that works for the largest number of people.” But that’s rarely the case with complex diseases, which is why Hussin’s team continues to sharpen its tools and plan for the oncoming wave of data. “We want to be ready to dissect those different levels of variability,” says Hussin.
Crédit photo : Julie Hussin


