COVID-19: Opening Up the Data
Pierre-Olivier Quirion is the switchboard operator at the centre of a consortium working to track COVID-19 strains in Quebec. This Rube-Golberg-like operation starts when a sample is swabbed from inside your nose and ends when the government releases its latest recommendations.
Quirion was also instrumental in making the data accessible to the general public as opposed to only making it available to other specialists. “Scientists do think about it,” he says over the phone. “But they’re already so busy, they aren’t under any obligations, and human and material resources to make it happen are scarce.”
The consortium—CoVSeQ—identified the 247 viral strains that led to the current outbreak in the province. Scientists from groups including the Institut national de santé publique du Québec, the McGill Genome Center and the Canadian Center for Computational Genomics (C3G), and Université de Montreal studied 734 viral samples from last February and March. They then compared them with nearly 22,000 samples from the rest of the world. The strains, they learned, entered with Quebecers returning from spring break trips abroad, and mostly originated in Europe and the Americas. The findings were released in a paper earlier this fall.
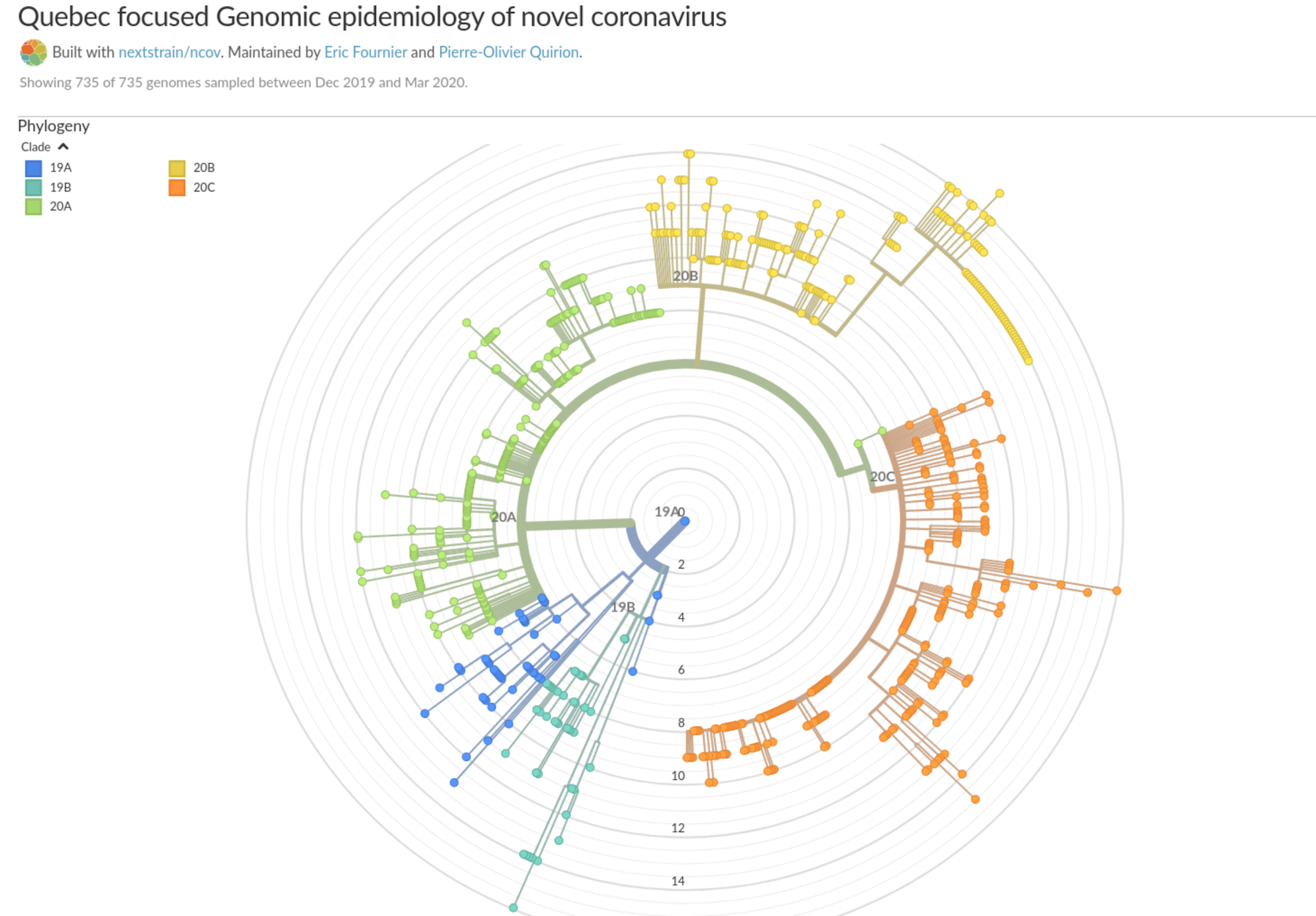
This image shows the 5 main branches of virus in Québec (from covseq.ca).
CoVSeQ’s main job is to find out which strains of SARS-CoV-2 are in the province. If they detect a new strain, they try to find out where it’s from. Public Health then applies measures to block possible entry points.
To identify different strains, which appear each time the virus mutates, genomic scientists sequence the viral samples. But you can’t just look at them. You first cut them into tiny pieces, then figure out how they fit together. “It’s like ripping a book apart and putting the pieces back in order,” says Quirion.
Phylogeneticists then compare the strains to determine which are most similar and probably have common ancestors. That’s how they know if someone returning from the Caribbean is carrying a strain that originated someplace else, like Europe.
And what’s Quirion’s role in this and why use supercomputers?
First, the coronavirus’s genome is long—it has about 30,000 ribonucleic acids, the basic molecules that make up genes. Second, you need to make many copies of each strain and repeat the process for better accuracy.
“[Researchers] often run their calculations on laptops,” Quirion notes, which wouldn’t work with the amount of data being crunched. “That’s where Calcul Québec comes into play,” he says. “It’s way more data than what’s usually analysed in such a short amount of time.”
Bioinformaticians at C3G develop and apply the software to piece together the genome and compare the strains. Working for both C3G and Calcul Québec, Quirion adapts the software to run on the latter’s powerful machines.
Genomics projects are the biggest users of Calcul Québec’s disk space. While physicists, astronomers and meteorologists run more complex calculations, the storage required for biological data is staggering. One of C3G’s projects alone uses 10% of Calcul Québec’s 25 million GB.
In this race to get results up the line, it’s easy to forget that, especially during a crisis, scientists have a responsibility to be transparent.
Like most scientists, CoVSeQ had every intention to publish their results to the scientific community. But there were no plans, at least initially, to make the data easily available to the public. Simply put, because there are no laws to enforce this practice, many organizations don’t dedicate resources towards it.
So Quirion insisted. He pushed to build a platform on which public data could be shared openly, and Calcul Québec agreed to facilitate it. The aim, as stated on CoVSeQ’s website, is “to provide interactive data visualizations to virologists, epidemiologists, public health officials and citizen scientists.” But it’s also there for journalists and any other citizen who wishes to see it.
Quirion explains that Calcul Québec has mainly been an accelerator in such projects, and a way for experts to work together. “I think Calcul Québec will play a much larger role in making data accessible to the public,” he concludes.

Pierre-Olivier ia an Advance Computing specialist. While he has a Phd in Physics, he now trades in Computational Biology at the Canadian Center for Computational Genetics and at Calcul Québec. He has honed his trade solving software problems in eclectic supercomputer environments, as an astrophysicist modeling stellar evolution and pulsation, as a Technical Director in animated film studios and as a professional in a Geriatric Hospital Neuroimaging lab.


